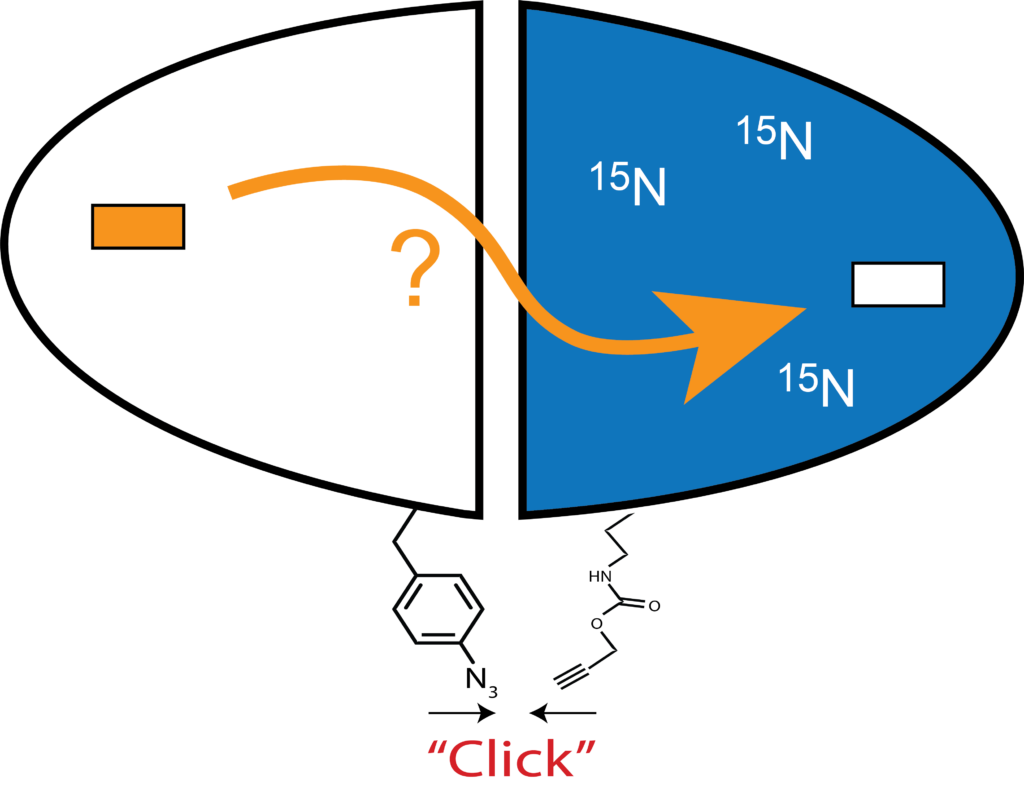
We are particularly attracted to homodimeric enzymes with allosteric properties. Two such enzymes under study in the lab are chorismate mutase (CM) and thymidylate synthase (TS). Symmetry is a classical feature of allosteric proteins (e.g. hemoglobin, aspartate transcarbamoylase, GroEL, etc.), but for NMR detection, signals from symmetric subunits cannot be distinguished. To address this problem, we construct “mixed labeled dimers” (MLDs), in which only one protomer (subunit) is labeled with NMR isotopes. An additional feature is to mutate the substrate binding site in one protomer, so that either the bound or empty subunit can be monitored by the vast number of NMR dynamics (“spin relaxation”) experiments can be performed. This gives a high-resolution picture of the structural and dynamic features of this singly liganded (“lig1”) allosteric intermediate, which is otherwise extremely difficult to study. The ability to observe the response of single protomers – especially the unbound – should yield fascinating, new insights into allosteric communication.
One problem with this approach is that subunits can exchange or “reapportion”. We stabilize the dimers to this exchange by chemically tethering the MLD subunits using biorthogonal “click chemistry”, which avoids use of cysteines (TS has active site cysteines), which enables them to be used for other experiments that utilize cysteine-based tags. Our approach uses the classic copper catalyzed azide-alkyne cycloaddition (CuAAC) reaction, and unique azide and alkyne groups are installed at specific sites using unnatural amino acids (UAAs) in combination with amber codon suppression methods.

We have found (Sapienza et al., below) that for CuAAC reactions with proteins in aqueous solution, efficient reactions are aided by removal of oxygen via a glove box.
Related Publications
Sapienza PJ, Currie MM, Lancaster NM, Li K, Aubé J, Goldfarb D, Cloer EW, Major MB, Lee AL, Visualizing an Allosteric Intermediate Using CuAAC Stabilization of an NMR Mixed Labeled Dimer, ACS Chem Biol (2021), 16(12):2766-2775. doi: 10.1021/acschembio.1c
