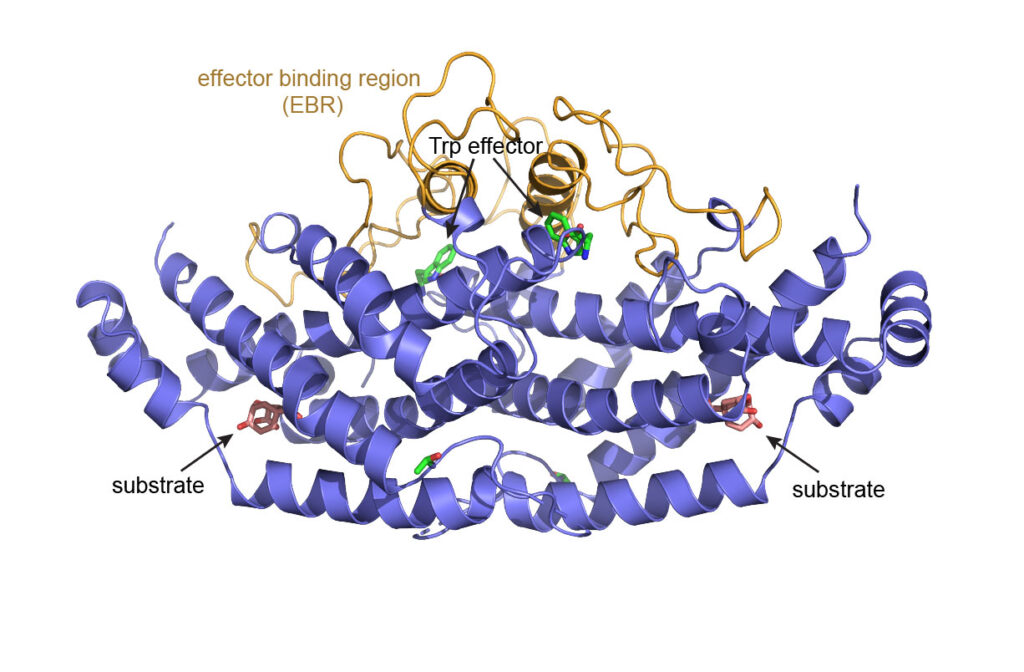
Chorismate mutase (CM) is a metabolic enzyme in the amino acid biosynthesis pathway that exhibits all the hallmarks of allostery. Yet, despite its larger size (60 kDa), it is amenable to detailed study by NMR. CM has a homodimeric structure that has been shown to adopt distinct, classical ‘tense’ (T) and ‘relaxed’ (R) quaternary conformations. CM reactivity is intrinsically positively cooperative, even though the two symmetric active sites are at opposite sites of the dimer, and CM can be both positively and negatively modulated by the allosteric effector ligands tryptophan and tyrosine. Application of NMR spectroscopy to this rich system should enable discovery of structural and dynamic processes that underlie classical allosteric regulation. In particular, we are interested in how the binding event in one protomer can extend to the other protomer active site – via conformational change or other dynamic processes – to stimulate higher activity.
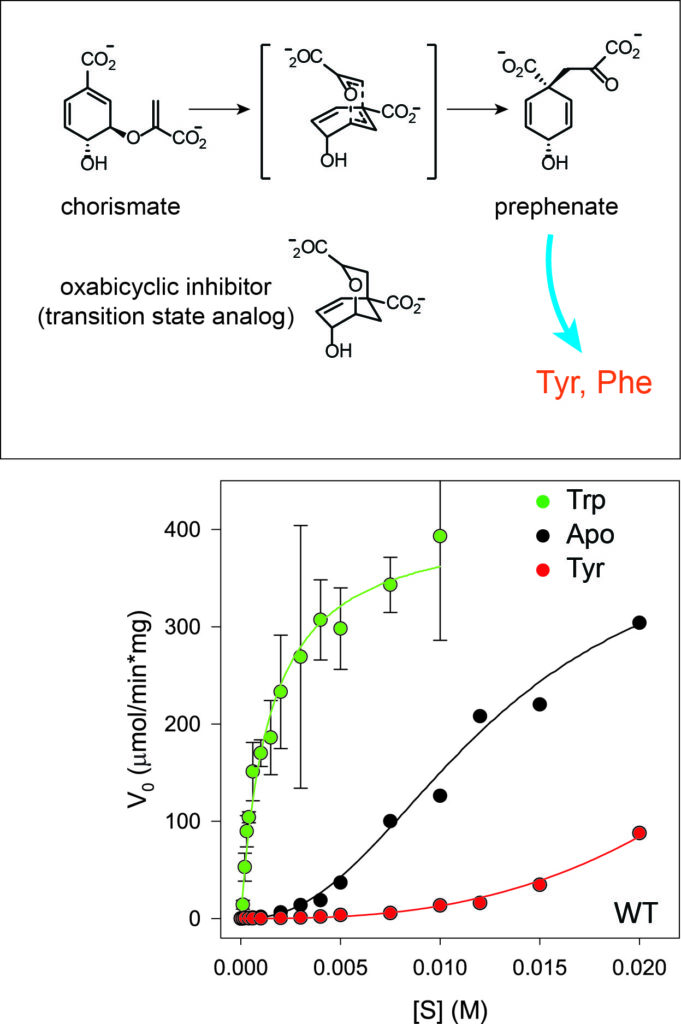
CM catalyzes the conversion of chorismic acid to prephenate, which is a precursor for synthesis of tyrosine and phenylalanine. Trp and Tyr amino acids bind to the same allosteric effector site, yet they have opposite effects on CM activity. In the absence of Trp/Tyr effectors, CM exhibits positive cooperativity, as evident from a sigmoidal velocity vs. [substrate] curve. The cooperativity is allosteric since the two active sites are separated by 40 Angstroms.
To aid the study of how CM responds to (substrate) binding in the active site, we collaborate with the Aube lab (CBMC Pharmacy) to synthesize a transition state analog compound (oxabicyclic inhibitor) to be used in experiments. The inhibitor compound binds with high affinity and is being used to 1) create mixed labeled dimers (MLDs), 2) drive CM into an ‘R’ conformation, and 3) measure allosteric cooperativity.
The CM system is ideal for structural, dynamic, and thermodynamic study of allostery. High-quality NMR spectra are obtained for the different allosteric states (apo, Trp-, Tyr-bound). We are currently using a battery of NMR relaxation, NMR titration, isothermal titration calorimetry (ITC), x-ray crystallography, and MD simulation approaches to reveal the underlying dynamic processes of allosteric communication and activity regulation.
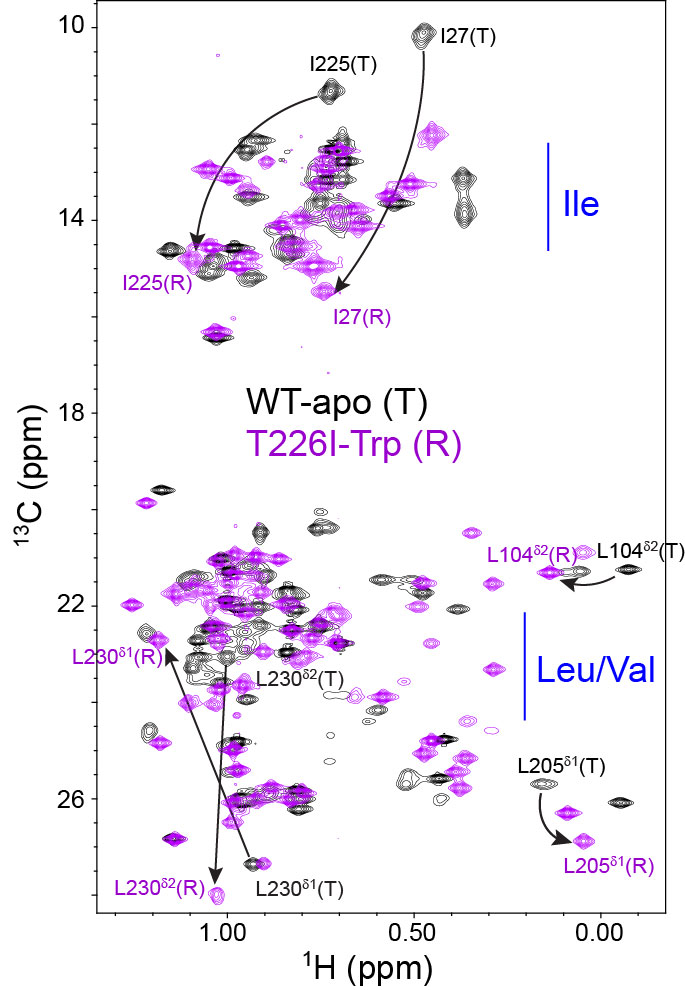
NMR spectra of methyl signals (contoured lines) from “tense” (T) and “relaxed” (R) allosteric conformational states of chorismate mutase (CM).
T: black R: purple
Methyl signals result from specific 1H and 13C labeling of isoleucine, leucine, and valine methyl groups against a deuterated background.
Large peak movements result upon the T-to-R allosteric conformational change.
Related Publications
Sapienza PJ, Bonin JP, Jinasena DHP, Li K, Dieckhaus H, Popov KI, Aubé J, and Lee AL, Mixed, non-classical behavior in a classic allosteric protein, Proc. Natl. Acad. Sci. 120, e2308338120 (2023).
