Overview
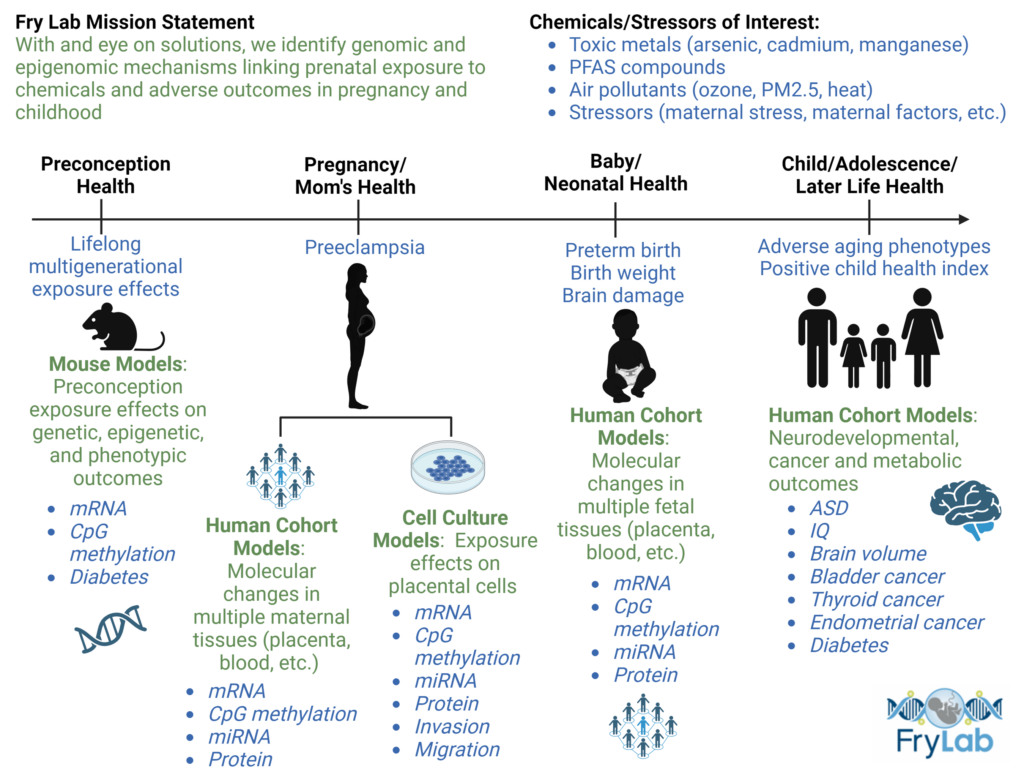
The Fry Lab research group studies how environmental exposures to toxic substances are associated with human disease with a particular focus on genomic and epigenomic perturbations. Epigenetics describes the subfield of genetics concerned with the alterations to DNA structure that modify gene expression without disturbing the underlying DNA sequence.These mechanisms can include DNA methylation, histone modification, and noncoding RNA.
Utilizing system biology and toxicogenomic perspectives, we identify key molecular pathways that associate environmental contaminants with adverse health outcomes. A current focus in the lab is maternal-child health, with interest in prenatal exposure to various toxic substances including arsenic, cadmium, and perflourinated chemicals. While exposures can occur in a variety of ways, this lab often examines exposures through drinking water. Drinking water quality is a major concern in North Carolina, with about three million residents relying on private wells and systems, which are not federally regulated. Our interdisciplinary team of researchers, conduct human population-based research, cell culture-based research and mouse model-based research to elucidate our understanding of exposures.
Cell Culture
Our cell culture team performs in vitro experiments using placental cell lines and primary placenta cells to learn how exposure to environmental toxicants affect gene expression, protein expression, epigenetic regulators, and cell migration. Some methods used to examine gene expression include reverse transcription polymerase chain reaction (RTPCR), mRNA nanostring sequencing, mRNA/miRNA sequencing, pyrosequencing, and the use of the EPIC 850k array. The team also employs scratch assays to investigate cell migration as well as western blots and ELISA assays to assess protein expression.
Human Population
Our human population team uses biostatistics, computational analyses, and epidemiological methods to relate chemical exposures in the environment to health outcomes. Multi-omic analyses further help in unraveling the underlying biology of environmental exposure-disease relationships. Additionally, the team examines geospatial trends and human biomonitoring to identify harmful exposures spanning North Carolina, the United States, and areas throughout the world.