Publications List
89. A. M. Solosky, I. M. Claudio, K. I. Kirkwood-Donelson, J. R. Chappel, M. G. Janech, F. M. Gullard, B. A. Neely ф, E. S. Baker ф, “Proteomic and Lipidomic Plasma Evaluations Reveal Biomarkers for the Diagnosis of Domoic Acid Toxicosis in California Sea Lions” J. Proteome Res. 2024.
88. E. S. Baker ф, H. Cooper ф, “2024 Emerging Investigators” J. Am. Soc. Mass Spectrom. 2024, 35, 2547-2553.
87. Y. V. Virkud, J. N. Styles, R. S. Kelly, S. U. Patil, B. Ruiter, N. P. Smith, C. Clish, C. E. Wheelock, J. C. Celedón, A. A. Litonjua, S. Bunyavanich, S. T. Weiss, E. S. Baker, J. A. Lasky-Su, W. G. Shreffler, “Immunomodulatory Metabolites in IgE-Mediated Food Allergy and Oral Immunotherapy Outcomes based on Metabolomic Profiling”. Pediatric Allergy and Immunology. 2024.
86. M. Giera, A. Aisporna, W. Uritboonthai, L. Hoang, R. Derks, K. M. Joseph, E. S. Baker, G. Siuzdak. “XCMS-METLIN: Data-Driven Metabolite, Lipid, and Chemical Analysis”. Mol. Syst. Biol. 2024.
85. A. S. McMillan*, G. Zhang*, M. K. Dougherty, S. K. McGill, A. S. Gulati, E. S. Baker, C. M. Theriot, “Metagenomic, Metabolomic, and Lipidomic Shifts Associated with Fecal Microbiota Transplantation in Clostridioides difficile Infection” mSphere. 2024.
84. J. R. Chappel, J. N. Dodds, K. I. Kirkwood Donelson, J. Fleming, D. M. Reif ф, E. S. Baker ф, “Streamlining Phenotype Classification and Highlighting Feature Candidates: A Screening Method for Non-Targeted Ion Mobility Spectrometry-Mass Spectrometry Data” Anal. Chem. 2024.
83. D. Kopczynski, C.S. Ejsing, J.G. McDonald, T. Bamba, E.S. Baker, J. Bertrand-Michel, B. Brügger, C. Coman, S.R. Ellis, T.J. Garrett, X. Li Guan, X. Han, M. Höring, M. Holčapek, N. Hoffmann, K. Huynh, R. Lehmann, J.W Jones, R. Kaddurah-Daouk, H.C. Köfeler, P.J. Meikle, T.O. Metz, V.B. O’Donnell, D. Saigusa, D. Schwudke, A. Shevchenko, F. Torta, R. Welti, M.R. Wenk, D. Wolrab, Y. Xia, K. Ekroos, R. Ahrends, G. Liebisch, “The Lipidomics Reporting Checklist: A Framework for Transparency of Lipidomic Experiments and Repurposing Resource Data” J. Lipid Res. 2024, 65.
82. E. C. Day, S. S. Chittari, K. C. Cunha, R. J. Zhao, J. N. Dodds, D. C. Davis, E. S. Baker, R. B. Berlow, J. E. Shea, R. U. Kulkarni, A. S. Knight, “A High-Throughput Workflow to Analyze Sequence-Conformation Relationships and Explore Hydrophobic Patterning in Disordered Peptides” Chem. 2024.
81. A. K. Boatman, J. R. Chappel, M. E. Polera, J. N. Dodds, S. M. Belcher, E. S. Baker, “Assessing Per- and Polyfluoroalkyl Substances (PFAS) in Fish Fillet using Non-Targeted Analyses” Environ. Sci. Technol. 2024.
80. J. N. Dodds, K. I. Kirkwood-Donelson, A. K. Boatman, D. R. U. Knappe, N. S. Hall, A. Schnetzer, E. S. Baker, “Evaluating Solid Phase Adsorption Toxin Tracking (SPATT) for Passive Monitoring of Per- and Polyfluoroalkyl Substances (PFAS) with Ion Mobility Spectrometry-Mass Spectrometry (IMS-MS)” Sci. Total Environ. 2024.
79. P. Qiao, M. T. Odenkirk, W. Zheng, Y. Wang, J. Chen, W. Xu, E. S. Baker, “Elucidating the Role of Lipid Interactions in Stabilizing the Membrane Protein KcsA” Biophys. J. 2024.
78. E. C. Day, K. C. Cunha, J. Zhao, A. J. DeStefano, J. N. Dodds, M. A. Yu, S. Han, E. S. Baker, J-E. Shea, R. Berlow, A. S. Knight, “Insights into the Conformational Ensembles of Compositionally Identical Disordered Peptidomimetics” Polym. Chem. 2024.
77. A. M. Solosky, K. I. Kirkwood-Donelson, M. T. Odenkirk, E. S. Baker, “Recent Additions and Access to a Multidimensional Lipidomic Database Containing Liquid Chromatography, Ion Mobility Spectrometry and Tandem Mass Spectrometry Information” Anal. Bioanal. Chem. 2024, ASAP.
76. R. A. Weed, G. Campbell, L. Brown, K. May, D. Sargent, E. Sutton, K. Burdette, W. Rider, E. S. Baker, J. Enders, “Non-Targeted PFAS Suspect Screening and Quantification of Drinking Water Samples Collected through Community Engaged Research in North Carolina’s Cape Fear River Basin” Toxics. 2024, 12, 403
75. E. S. Baker, W. Uritboonthai, A. Aisporna, C. Hoang, H. M. Heyman, M. Giera, G. Siuzdak, “METLIN-CCS Lipid Database: An Authentic Standards Resource for Lipid Classification and Identification” Nat. Metab. 2024, ASAP.
74. M. T. Odenkirk, X. Zheng, J. E. Kyle, K. G. Stratton, C. D. Nicora, K. Bloodsworth, C. A. Mclean, C. L. Masters, M. E. Monroe, J. D. Doecke, R. D. Smith, K. E. Burnum-Johnson, B. Roberts, E. S. Baker, “Deciphering ApoE Genotype-Driven Proteomic and Lipidomic Alternations in Alzheimer’s Disease Across Distinct Brain Regions” J. Proteome Res. 2024, 23, 2970.
73. K. I. Kirkwood-Donelson, J. Chappel, E. Tobin, J. N. Dodds, D. M. Reif, J. C. DeWitt, E. S. Baker, Investigating Mouse Hepatic Lipidome Dysregulation Following Exposure to Emerging Per- and Polyfluoroalkyl Substances (PFAS). Chemosphere, 2024, 384, 141654. https://doi.org/10.1016/j.chemosphere.2024.141654
72. J. F. Fleming, J. S. House, J. R. Chappel, A. A. Motsinger-Reif, D. M. Reif, “Guided optimization of ToxPi model weights using a Semi-Automated approach” Computational Toxicology, Volume 29, 2024, ISSN 2468-1113, https://doi.org/10.1016/j.comtox.2023.100294.
71. E. C. Gentry, S. L. Collins, M. Panitchpakdi, P. Belda-Ferre, A. K. Stewart, M. C. Terrazas, H-H. Lu, S. Zuffa, T. Yan, J. Avila-Pacheco, D. R. Plichta, A T. Aron, M. Wang, A. K. Jarmusch, F. Hao, M. Syrkin-Nikolau, H. Vlamakis, A. N. Ananthakrishnan, B. Boland, A. Hemperly, N. V. Casteele, F. J. Gonzalez, C. B. Clish, R. J. Xavier, H. Chu, E. S. Baker, A. D. Patterson, R. Knight, D. Siegel, P. C. Dorrestein, “Reverse metabolomics for the discovery of chemical structures from humans” Nat. 2024, 626, 419-426. https://doi.org/10.1038/s41586-023-06906-8
70. S. K. Witchey, M. G. Doyle, J. D. Fredenburg, G. St Armour, B. Horman, M. T. Odenkirk, D. L.
Aylor, E. S. Baker, H. B. Patisaul, “Impacts of Gestational FireMaster 550 (FM 550) Exposure on the
Neonatal Cortex are Sex Specific and Largely Attributable to the Organophosphate Esters”
Neuroendocrinology. 2023, 113, 1262-1282.
69. J. P. Ryan, M. Kostelic, C-C. Hsieh, J. Powers, C. Aspinwall, J. Schiel, M. T. Marty, E. S. Baker, “Characterizing Adeno-Associated Virus Capsids with Both Denaturing and Intact Analysis Methods” J. Am. Soc. Mass Spectrom. 2023, 34, 2811-2821. https://doi.org/10.1021/jasms.3c00321
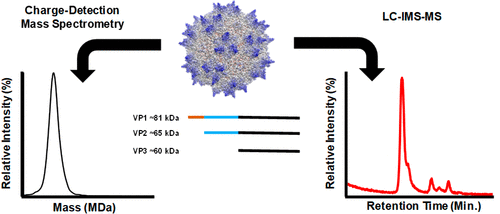
68. E. S. Baker, C. Hoang, W. Uritboonthai, H. M. Heyman, B. Pratt, M. MacCoss, B. MacLean, R. Plumb, A. Aisporna, G. Siuzdak, “METLIN-CCS: An Ion Mobility Spectrometry Collision Cross Section Database” Nat. Methods. 2023, 20, 1836-1837. https://doi.org/10.1038/s41592-023-02078-5.
67. H. Cooper, E. S. Baker, “2023 Emerging Investigators” J. Am. Soc. Mass Spectrom. 2023, 34, 1821-1825.
66. K. I. Kirkwood-Donelson, J. N. Dodds, A. Schnetzer, N. Hall, E. S. Baker, “Uncovering per- and polyfluoroalkyl substances (PFAS) with nontargeted ion mobility spectrometry–mass spectrometry analyses.” Sci. Adv. 9, eadj7048 (2023). DOI:10.1126/sciadv.adj7048.
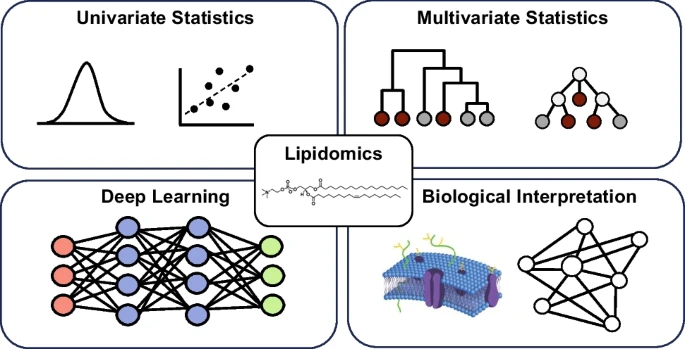
65. J. R. Chappel, K. I. Kirkwood-Donelson, D. M. Reif, E. S. Baker, “From big data to big insights: statistical and bioinformatic approaches for exploring the lipidome” Anal Bioanal Chem (2023). https://doi.org/10.1007/s00216-023-04991-2.
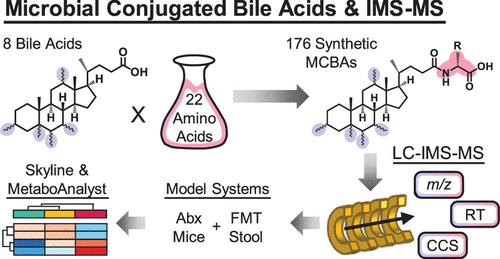
64. A. K. Stewart, M. H. Foley, M. K. Dougherty, S. K. McGill, A. S. Gulati, E. C. Gentry, L. R. Hagey, P. C. Dorrestein, C. M. Theriot, J. N. Dodds, E. S. Baker, “Using Multidimensional Separations to Distinguish Isomeric Amino Acid–Bile Acid Conjugates and Assess Their Presence and Perturbations in Model Systems” Anal. Chem. 2023, 95, 15357-15366. DOI: 10.1021/acs.analchem.3c03057.
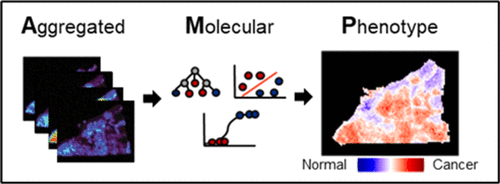
63. J. R. Chappel, M. E. King, J. Fleming, L. S. Eberlin, D. M. Reif, E. S. Baker, “Aggregated Molecular Phenotype Scores: Enhancing Assessment and Visualization of Mass Spectrometry Imaging Data for Tissue-Based Diagnostics” Anal. Chem. 2023, 95, 12913-12922. DOI: 10.1021/acs.analchem.3c02389.
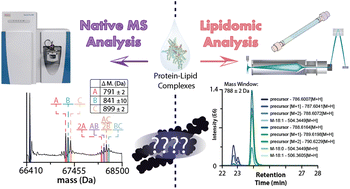
62. Y. Zhu, M. T. Odenkirk, P. Qao*, T. Zhang, S. Schrecke, M. Zhou, M. T. Marty, E. S. Baker, A. Laganowsky. Combining Native Mass Spectrometry and Lipidomics to Uncover Specific Membrane Protein-lipid Interactions from Natural Lipid Sources. Chem. Sci. 2023, 14, 8570-8582.
61. J. C. Tran, I. D. G. Campuzano, E. S. Baker, V. H. Wysocki, “Introducing the High-Throughput in
Mass Spectrometry Special Focus Issue” J. Am. Soc. Mass Spectrom. 2023, 34, 1543-1544.
60. A. C. Cordova, W. D. Klaren, L. C. Ford, F. A. Grimm, E. S. Baker, Y.-H. Zhou, F. A. Wright, I.
Rusyn, “Integrative Chemical-Biological Grouping of Complex High Production Volume Substances
from Lower Olefin Manufacturing Streams” Toxics. 2023, 11, 586.
59. E. S. Baker, “Assessing How Chemical Exposures Affect Human Health” LCGC. 2023, 36, 7-10.
58. A. Bilbao, N. Munoz, J. Kim, D. J. Orton, Y. Gao, K. Poorey, K. Pomraning, K. Weitz, M. Burnet, C.
D. Nicora, R. Wilton, S. Deng, Z. Dai, E. Oksen, A. Gee, F. A. Fasani, A. Tsalenko, D. Tanjore, J.
Gardner, R. D. Smith, J. K. Michener, J. M. Gladden, E. S. Baker, C. J. Petzold, Y-M. Kim, A.
Apffel, J. K. Magnuson, K. E. Burnum-Johnson, “PeakDecoder Enables Machine Learning-based
Metabolite Annotation and Accurate Profiling in Multidimensional Mass Spectrometry
Measurements” Nat. Commun. 2023, 14, 2461.
57. K. I. Kirkwood, M. T. Odenkirk, E. S. Baker, “Ion Mobility Spectrometry” In: Holčapek M, Ekroos K (eds) Mass Spectrometry for Lipidomics: Methods and Applications. Wiley (2023).
56. M. H. Foley, M. E. Walker, A. K. Stewart; S. O’Flaherty, E. C. Gentry, S. Patel, V. V. Beaty, G. Allen, M. Pan, J. B. Simpson, C. Perkins, M. E. Vanhoy, M. K. Dougherty, S. K. McGill, A. S. Gulati, P. C. Dorrenstein, E. S. Baker, M. R. Redinbo, R. Barrangou, C. M. Theriot, “Bile Salt Hydrolases Shape the Bile Acid Landscape and Restrict Clostridioides difficile Growth in the Murine Gut.” Nat. Microbiol. 2023.
55. R. A. Weed, A. K. Boatman, J. R. Enders, “Recovery of Per- and Polyfluoroalkyl Substances After Solvent Evaporation” Environ. Sci.: Processes Impacts 2022, 24 (12), 2263-2271.
54. M. A. Rainey, C. A. Watson, C. K. Asef, M. R. Foster, E. S. Baker, F. M. Fernández, “CCS
Predictor 2.0: An Open-Source Jupyter Notebook Tool for Filtering Out False Positives in
Metabolomics” Anal. Chem. 2022, 94, 17456-17466.
53. A. T. Roman-Hubers, C. Aeppli, J. N. Dodds, E. S. Baker, K. M. McFarlin, D. J. Letinski, L. Zhao,
D. A. Mitchell, T. F. Parkerton, R. C. Prince, T. Nedwed, I. Rusyn, “Temporal Chemical Composition
Changes in Water Below a Crude Oil Slick Irradiated with Natural Sunlight” Marine Pollution
Bulletin 2022, 185, 114360.
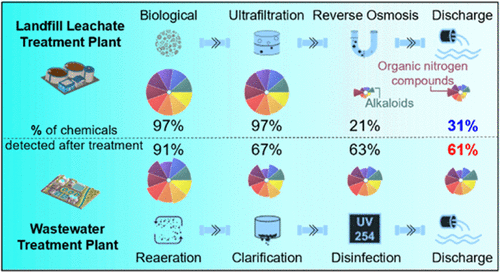
52. M. G. Doyle, M. T. Odenkirk, A. K. Stewart, J. P. Nelson, E. S. Baker, and F. De La Cruz “Assessing the Fate of Dissolved Organic Compounds in Landfill Leachate and Wastewater Treatment Systems”. ACS EST Water. 2022, 2(12), 2502-2509.
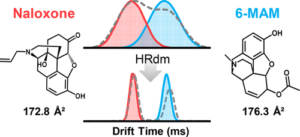
51. K. E. Butler and E. S. Baker “A High-Throughput Ion Mobility Spectrometry Mass Spectrometry Screening Method for Opioid Profiling“. J. Am. Soc. Mass Spectrom. 2022, 33(10), 1904-1913.
50. A. Valdiviezo, Y. Kato, E. S. Baker, W. A. Chiu, I. Rusyn, “Evaluation of Metabolism of a Defined
Pesticide Mixture Through Multiple In Vitro Liver Models” Toxics. 2022, 10, 566.
49. M. M. Kostelic, J. P. Ryan, L. S. Brown, T. W. Jackson, C. C. Hsieh, C. K. Zak, H. M. Sanders, Y. Liu, V. Shugui Chen, M. Byrne, C. A. Aspinwall, E. S. Baker, and M. T. Marty “Stability and Dissociation of Adeno-Associated Viral Capsids by Variable Temperature-Charge Detection-Mass Spectrometry” Anal. Chem. 2022, 94(34), 11723-11727.
48. J. G. McDonald, C. S. Ejsing, D. Kopczynski, M. Holčapek, J. Aoki, M. Arita,M. Arita, E. S. Baker,
J. Bertrand-Michel, J. A. Bowden, B. Brügger, S. Ellis, M. Fedorova, W. J. Griffiths, X. Han, J.
Hartler, N. Hoffman, J. P. Koelmel, H. C. Köfeler, T. W. Mitchell, V. B. O’Donnell, D. Saigusa, D.
Schwudke, A. Shevchenko, C. Z. Ulmer, M. R. Wenk, M. Witting, D. Wolrab, Y. Xia, R. Ahrends, G.
Liebisch, K. Ekroos, “Introducing the Lipidomics Minimal Reporting Checklist” Nat. Metab. 2022, 4,
1086-1088.
47. K I. Kirkwood, B. S. Pratt, N. Shulman, K. Tamura, M.l J. MacCoss, B. X. MacLean, E. S. Baker “Utilizing Skyline to analyze lipidomics data containing liquid chromatography, ion mobility spectrometry and mass spectrometry dimensions” Nat. Protoc. 2022, 17, 3415-2430.
46. M.Foster, M. Rainey, C. Watson, J. N. Dodds, K. I. Kirkwood, F. M. Fernández, E. S. Baker “Uncovering PFAS and Other Xenobiotics in the Dark Metabolome Using Ion Mobility Spectrometry, Mass Defect Analysis, and Machine Learning” Environ. Sci. Technol. 2022, 56(12), 9133-9143.
45. M. M. Kostelic, C. C. Hsieh, H. M. Sanders, C. K. Zak, J. P. Ryan, E. S. Baker, C. A. Aspinwall, M. T. Marty “Surface Modified Nano-Electrospray Needles Improve Sensitivity for Native Mass Spectrometry” J. Am. Soc. Mass Spectrom. 2022, 33(6), 1031-1037.
44. Y. Zhu, S. Schrecke, S.Tang, M. T. Odenkirk, T. Walker, L. Stover, J. Lyu, T. Zhang, D. Russell, E. S. Baker, X. Yan, A. Laganowsky, “Cupric Ions Selectively Modulate TRAAK-Phosphatidylserine Interactions” J. Am. Chem. Soc. 2022, 144(16), 7048-7053.
43. K. E. Butler, J. N. Dodds, T. Flick, I. D. G. Campuzano, E. S. Baker “High Resolution Demultiplexing (HRdm) Ion Mobility Spectrometry-Mass Spectrometry for Aspartic and Isoaspartic Acid Determination and Screening” Anal. Chem. 2022, 94(16), 6191-6199.
42. A. Bilbao, B. C. Gibbons, S. Stow, J. E. Kyle, K. J. Bloodsworth, S. H. Payne, R. D. Smith, Y. M.
Ibrahim, E. S. Baker, J. C. Fjeldsted, “A Preprocessing Tool for Enhanced Ion Mobility-Mass
Spectrometry-Based Omics Workflows” J. Proteome Res. 2022, 21, 798-807.
41. K. I. Kirkwood, J. Fleming, H. Nguyen, D. M. Reif, E. S. Baker, S. M. Belcher “Utilizing Pine Needles to Temporally and Spatially Profile Per- and Polyfluoroalkyl Substances (PFAS)” Environ. Sci. Technol. 2022 56(6), 3441-3451.
40. A. T. Hubers-Roman, A. C. Cordova, A. M. Rohde, W. A. Chiu, T. J. McDonald, F. A. Wright, J. N. Dodds, E. S. Baker, I. Rusyn, “Characterization of compositional variability in petroleum substances” Fuel 2022, 317(1), 123547.
39. E. Rampler, E. S. Baker, K. I. Kirkwood, M. Schwaiger-Haber, M. Tam, M. A. Jones, M. Sherman, “Empowering Women and Addressing Underrepresentation in the Field of Mass Spectrometry“ Expert Rev. Proteom. 2022 19(1), 1-3.
38. J. N. Dodds, L. Wang, G. J. Patti, E. S. Baker, “Combining Isotopologue Workflows and Simultaneous Multidimensional Separations to Detect, Identify, and Validate Metabolites in Untargeted Analyses” Anal. Chem. 2022 94(5), 2527-2535.
37. E. S. Baker , D. R. U. Knappe, “Per- and Polyfluoroalkyl Substances (PFAS) – Contaminants of
Emerging Concern” Anal. Bioanal. Chem. 2022, 414, 1187-1188.
36. K. I. Kirkwood, M. W. Christopher, J. L. Burgess, S. R. Littau, K. Foster, K. Richey, B. S. Pratt, N. Shulman, K. Tamura, M. J. MacCoss, B. X. MacLean, E. S. Baker “Development and Application of Multidimensional Lipid Libraries to Investigate Lipidomic Dysregulation Related to Smoke Inhalation Injury Severity” J. Prot. Res. 2022 21(1), 232-242.
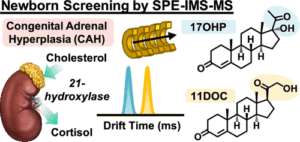
35. J. N. Dodds, E. S. Baker “Improving the Speed and Selectivity of Newborn Screening Using Ion Mobility Spectrometry-Mass Spectrometry” Anal Chem. 2021, 95(51), 17094-17102.
34. A. Valdiviezo, N. A. Aly, Y. Luo, A. Cordova, G. Casillas, M. Foster, E. S. Baker, I. Rusyn, “Analysis of Per- and Polyfluoroalkyl Substances in Houston Ship Channel and Galveston Bay Following a Large-scale Industrial Fire Using Ion Mobility Spectrometry-Mass Spectrometry” J. Environ. Sci. 2022, 115, 350-362.
33. M. T. Odenkirk, B. M. Horman, J. N. Dodds, H. B. Patisaul, E. S. Baker “Combining Micropunch Histology and Multidimensional Lipidomic Measurements for In-Depth Tissue Mapping” ACS Meas. Sci. Au 2022, 2(1), 67-75.
32. N. A. Aly, J. N. Dodds, Y. S. Luo, F. A. Grimm, M. Foster, Ivan Rusyn, E. S. Baker “Utilizing Ion Mobility Spectrometry-Mass Spectrometry for the Characterization and Detection of Persistent Organic Pollutants and Their Metabolites.” Anal. Bioanal. Chem. 2021.
31. K. E. Butler, J. G. Kalmar, D. C. Muddiman, E. S. Baker “Utilizing Liquid Chromatography, Ion Mobility Spectrometry, and Mass Spectrometry to Assess INLIGHT Derivatized N-linked Glycans in Biological Samples” Anal. Bioanal. Chem. 2021, s00216.
30. D. Zamith-Miranda, H. M. Heyman, M. C. Burnet, S. P. Couvillion, X. Zheng, N. Munoz, J. E. Kyle,
E. M. Zink, K. K. Weitz, K. J. Bloodsworth, G. Clair, J. D. Zucker, J. R. Teuton, S. H. Payne, Y.-M.
Kim, M. R. Gil, E. S. Baker, E. L. Bredeweg, J. D. Nosanchuk, E. S. Nakayasu, “A Histoplasma
capsulatum Lipid Metabolic Map Identifies Antifungal Targets” mBio 2021, 12, e02972-21.
29. K. D. Duncan, X. Sun, E. S. Baker, S. K. Dey, I. Lanekoff, “In Situ Imaging Reveals Disparity
Between Prostaglandin Localization and Abundance of Prostaglandin Synthases” Commun. Biol.
2021, 4, 1-9.
28. A. T. Roman-Hubers, A. Cordova, N. A. Aly, T. J. McDonald, D. Lloyd, F. A. Wright, E. S. Baker,
W. A. Chiu, I. Rusyn, “A Data Processing Workflow to Identify Structurally Related Compounds in
Petroleum Substances Using Ion Mobility Spectrometry-Mass Spectrometry” Energy Fuels. 2021, 40,
1034-1049.
27. G. Sansom, K. Kirsch, G. Casillas, K. Camargo, T. L. Wade, A. H. Knap, E. S. Baker, J. A. Horney, “Spatial Distributions of Polycyclic Aromatic Hydrocarbon Contaminants after Hurricane Harvey in a Houston Neighborhood” J. Health and Pollution. 2021, 11, 210308.
26. M. R. Ligare, K. A. Morrison, M. A. Hewitt, J. U. Reveles, N. Govind, H. Hernandez, E. S. Baker,
B. H. Clowers, J. Laskin, G. E. Johnson, “Ion Mobility Spectrometry Characterization of the
Intermediate Hydrogen-Containing Gold Cluster Au 7 (PPh 3 ) 7 H 5 2+ ” J. Phys. Chem. Lett. 2021, 12, 2502-2508.
25. M. T. Odenkirk, D. M. Reif, E. S. Baker “Multiomic Big Data Analysis Challenges: Increasing Confidence in the Interpretation of Artificial Intelligence Assessments” Anal Chem. 2021, 0c04850.
24. L. Khadempour, J. E. Kyle, B. M. Webb-Robertson, C. D. Nicora, F. B. Smith, R. D. Smith, M. S. Lipton, C. R. Currie, E. S. Baker, K. E. Burnum-Johnson, “From Plants to Ants: Fungal Modification of Leaf Lipids for Nutrition and Communication in the Leaf-Cutter Ant Fungal Garden Ecosystem” mSystems 2021, 6(2), e01307-20
23. K. E. Butler, Y. Takinami, A. Rainczuk, E. S. Baker, B. R. Roberts “Utilizing Ion Mobility-Mass Spectrometry to Investigate the Unfolding Pathway of Cu/Zn Superoxide Dismutase” Front. Chem. 2021, 9.
22. Y-S. Luo, Z. Chen, A. D. Blanchette, Y. H. Zhou, F. A. Wright, E. S. Baker, W. A. Chiu, I. Rusyn,
“Relationships Between Constituents of Energy Drinks and Beating Parameters in Human Induced
Pluripotent Cell (iPSC)-Derived Cardiomyocytes” Food Chem. Toxicol. 2021, 149, 111979.
21. M. T. Odenkirk, K. G. Stratton, L. M. Bramer, B. J. M. Webb-Robertson, K. J. Bloodsworth, M. E. Monroe, K. E. Burnum-Johnson, E. S. Baker “From Prevention to Disease Perturbations: A Multi-Omic Assessment of Exercise and Myocardial Infarctions.” Biomolecules 2021, 11, 40.
20. H. C. Kofeler, R. Ahrends, E. S. Baker, K. Ekroos, X. Han, N. Hoffmann, M. Holcapek, M. R. Wenk, G. Liebisc, “Recommendations for good practice in MS-based lipidomics.” J. Lipid Res. 2021, 62, 100138.
19. A. T. Roman‐Hubers, T. J. McDonald, E. S. Baker, W. A. Chiu, I. Rusyn, “A comparative analysis of analytical techniques for rapid oil spill identification.” Environ. Toxicol. Chem. 2021, 40, 1034-1049.
18. J. N. Dodds, N. L. M. Alexander, K. I. Kirkwood, M. R. Foster, Z. R. Hopkins, D. R. U. Knappe, E. S. Baker, “From Pesticides to PFAS: An Evaluation of Recent Targeted and Untargeted Mass Spectrometry Methods for Xenobiotics.” Anal. Chem. 2020, 93(1), 641-656.
17. Y. S. Luo, N. A. Aly, J. McCord, M. J. Strynar, W. A. Chiu, J. N. Dodds, E. S. Baker, I. Rusyn, “Rapid Characterization of Emerging Per- and Polyfluoroalkyl Substances (PFAS) in Aqueous Film-Forming Foams (AFFF) Using Ion Mobility Spectrometry-Mass Spectrometry (IMS-MS).“ Environ. Sci. Technol. 2020, 54(23), 15024-15034.
16. E. S. Orwoll, J. Wiedrick, C.M. Nielson, J. Jacobs, E. S. Baker, P. Piehowski, V. Petyuk, Y. Gao, T. Shi, R. D. Smith, D. C. Bauer, S. R. Cummings, J. Lapidus, “Proteomic assessment of serum biomarkers of longevity in older men.” Aging Cell 2020, 19(11).
15. M. T. Odenkirk, P. P. K. Zin, J. R. Ash, D. M. Reif, D. Fourches, E. S. Baker, “Structural-based connectivity and omic phenotype evaluations (SCOPE): A cheminformatics toolbox for investigating lipidomic changes in complex systems.” Analyst 2020, 145, 7197-7209.
14. M. T. Odenkirk, K. G. Stratton, M. A. Gritsenko, L. M. Bramer, B. J. M. Webb-Robertson, K. J. Bloodsworth, K. K. Weitz, A. K. Lipton, M. E. Monroe, J. R. Ash, D. Fourches, B. D. Taylor, K. E. Burnum-Johnson, E. S. Baker, “Unveiling molecular signatures of preeclampsia and gestational diabetes mellitus with multi-omics and innovative cheminformatics visualization tools.” Mol. Omics. 2020, 16(6), 521-532.
13. J. G. Kalmar, K. E. Butler, E. S. Baker, D. C. Muddiman, “Enhanced protocol for quantitative N-linked glycomics analysis using Individuality Normalization when Labeling with Isotopic Glycan Hydrazide Tags (INLIGHT).” Anal. Bioanal. Chem. 2020, 412, 7569-7579.
12. N. A. Aly, Y. S. Luo, Y. Liu, G. Casillas, T. J. McDonald, J. M. Kaihatu, M. Jun, N. Ellis, S. Gossett, J. N. Dodds, E. S. Baker, S. Bhandari, W. A. Chiu, I. Rusyn, “Temporal and spatial analysis of per and polyfluoroalkyl substances in surface waters of Houston ship channel following a large-scale industrial fire incident.” Environ. Pollut. 2020, 265B.
11. M. T. Soper-Hopper, J. Vandegrift, E. S. Baker, F. Fernandez, “Metabolite collision cross section prediction without energy-minimized structures.” Analyst, 2020, 145, 5414-5418.
10. J. N. Dodds, Z. R. Hopkins, D. R. U. Knappe, E. S. Baker, “Rapid Characterization of Per- and Polyfluoroalkyl Substances (PFAS) by Ion Mobility Spectrometry-Mass Spectrometry (IMS-MS)” Anal. Chem. 2020, 92(6), 4427-4435.
9. M. Ekelof, J. Dodds, S. Khodjaniyazova, K. P. Garrard, E. S. Baker, D. C. Muddiman, “Coupling IR-MALDESI with Drift Tube Ion Mobility-Mass Spectrometry for High-Throughput Screening and Imaging Applications” J. Am. Soc. Mass Spectrom. 2020, 31(3), 642-650.
8. M. T. Odenkirk, E. S. Baker, “Utilizing Drift Tube Ion Mobility Spectrometry for the Evaluation of Metabolites and Xenobiotics. In: Paglia G., Astarita G. (eds) Ion Mobility-Mass Spectrometry” Methods in Molecular Biology. 2020, vol 2084. Humana, New York, NY.
7. Y. Zhang, Y. Zhong, A. Connor, D. Miller, R. Cao, J. Shen, B. Song, E. S. Baker, Q. Tang, S.
Pulavarti, R. Liu, Q. Wang, Z. Lu, T. Szyperski, H. Zeng, X. Li, R. D. Smith, E. Zureck, J. Zhu, B.
Gong, “Folding and Assembly of Short α, β, γ-Hybrid Peptides: Minor Variations in Sequence and
Drastic Differences in Higher-Level Structures” J. Am. Chem. Soc. 2019, 141, 14239-14248.
6. J. N. Dodds, E. S. Baker, “Ion Mobility Spectrometry: Fundamental Concepts, Instrumentation, Applications, and the Road Ahead” J. Am. Soc. Mass Spectrom. 2019, 30(11), 2185-2195.
5. E. S. Baker, G. J. Patti, “Perspectives on Data Analysis in Metabolomics: Points of Agreement
and Disagreement from the ASMS Fall Workshop” J. Am. Soc. Mass Spectrom. 2019, 30, 2031-2036.
4. X. Zheng, F. B. Smith, N. A. Aly, J. Cai, R. D. Smith, A. D. Patterson, E. S. Baker, “Evaluating the Structural Complexity of Isomeric Bile Acids with Ion Mobility Spectrometry” Anal. Bioanal. Chem. 2019, 411, 4673–4682.
3. K. E. Burnum-Johnson, X. Zheng, J. N. Dodds, J. Ash, D. Fourches, C. D. Nicora, J. P. Wendler, T. O. Metz, K. M Waters, J. K. Jansson, R. D. Smith, E. S. Baker, “Ion Mobility Spectrometry and the Omics: Distinguishing Isomers, Isobars, Molecular Classes and Contaminant Ions in Complex Samples” Trends Anal. Chem. 2019, 116, 292-299.
2. P. L. Plante, E. Francovic-Fontain, J. C. May, J. A. McLean, E. S. Baker, F. Laviolette, M. Marchard, J. Corbeil, “Predicting Ion Mobility Collision Cross Sections Using a Deep Neural Network: DeepCCS” Anal. Chem. 2019, 91, 5191-5199.
1. M. E. Monge, J.N. Dodds, E. S. Baker, A. S. Edison, F. M. Fernández, “Challenges in Identifying the Dark Molecules of Life” Annu. Rev. Anal. Chem. 2019, 12, 177-199.
Dr. Baker’s publications prior to NCSU can be found on Google Scholar.